publications
filter by type:
Quantification of Nematic Cell Polarity in Three-dimensional Tissues
How epithelial cells coordinate their polarity to form functional tissues is an open question in cell biology. Here, we characterize a unique type of polarity found in liver tissue, nematic cell polarity, which is different from vectorial cell polarity in simple, sheet-like epithelia. We propose a conceptual and algorithmic framework to characterize complex patterns of polarity proteins on the surface of a cell in terms of a multipole expansion. To rigorously quantify previously observed tissue-level patterns of nematic cell polarity (Morales-Navarette et al., eLife 2019), we introduce the concept of co-orientational order parameters, which generalize the known biaxial order parameters of the theory of liquid crystals. Applying these concepts to three-dimensional reconstructions of single cells from high-resolution imaging data of mouse liver tissue, we show that the axes of nematic cell polarity of hepatocytes exhibit local coordination and are aligned with the biaxially anisotropic sinusoidal network for blood transport. Our study characterizes liver tissue as a biological example of a biaxial liquid crystal. The general methodology developed here could be applied to other tissues and in-vitro organoids.
PLoS Computational Biology,
2020
An interpretable automated detection system for FISH-basedHER2 oncogene amplification testing in histo-pathologicalroutine images of breast and gastric cancer diagnostics
Histo-pathological diagnostics are an inherent part of the everyday work but are particularly laborious and associated with time-consuming manual analysis of image data. In order to cope with the increasing diagnostic case numbers due to the current growth and demographic change of the global population and the progress in personalized medicine, pathologists ask for assistance. Profiting from digital pathology and the use of artificial intelligence, individual solutions can be offered (e.g. detect labeled cancer tissue sections). The testing of the human epidermal growth factor receptor 2 (HER2) oncogene amplification status via fluorescence in situ hybridization (FISH) is recommended for breast and gastric cancer diagnostics and is regularly performed at clinics. Here, we develop an interpretable, deep learning (DL)-based pipeline which automates the evaluation of FISH images with respect to HER2 gene amplification testing. It mimics the pathological assessment and relies on the detection and localization of interphase nuclei based on instance segmentation networks. Furthermore, it localizes and classifies fluorescence signals within each nucleus with the help of image classification and object detection convolutional neural networks (CNNs). Finally, the pipeline classifies the whole image regarding its HER2 amplification status. The visualization of pixels on which the networks’ decision occurs, complements an essential part to enable interpretability by pathologists.
MIDL 2020 (medical imaging with deep learming),
2020
Haematopoietic stem cells in perisinusoidal niches are protected from ageing
With ageing, intrinsic haematopoietic stem cell (HSC) activity decreases, resulting in impaired tissue homeostasis, reduced engraftment following transplantation and increased susceptibility to diseases. However, whether ageing also affects the HSC niche, and thereby impairs its capacity to support HSC function, is still widely debated. Here, by using in-vivo long-term label-retention assays we demonstrate that aged label-retaining HSCs, which are, in old mice, the most quiescent HSC subpopulation with the highest regenerative capacity and cellular polarity, reside predominantly in perisinusoidal niches. Furthermore, we demonstrate that sinusoidal niches are uniquely preserved in shape, morphology and number on ageing. Finally, we show that myeloablative chemotherapy can selectively disrupt aged sinusoidal niches in the long term, which is linked to the lack of recov-ery of endothelial Jag2 at sinusoids. Overall, our data characterize the functional alterations of the aged HSC niche and unveil that perisinusoidal niches are uniquely preserved and thereby protect HSCs from ageing.
Nature Cell Biology,
2019
Liquid-crystal organization of liver tissue
Functional tissue architecture originates by self-assembly of distinct cell types, following tissue-specific rules of cell-cell interactions. In the liver, a structural model of the lobule was pioneered by Elias in 1949. This model, however, is in contrast with the apparent random 3D arrangement of hepatocytes. Since then, no significant progress has been made to derive the organizing principles of liver tissue. To solve this outstanding problem, we computationally reconstructed 3D tissue geometry from microscopy images of mouse liver tissue and analyzed it applying soft-condensed-matter-physics concepts. Surprisingly, analysis of the spatial organization of cell polarity revealed that hepatocytes are not randomly oriented but follow a long-range liquid-crystal order. This does not depend exclusively on hepatocytes receiving instructive signals by endothelial cells, since silencing Integrin-β1 disrupted both liquid-crystal order and organization of the sinusoidal network. Our results suggest that bi-directional communication between hepatocytes and sinusoids underlies the self-organization of liver tissue.
eLIFE 2019;8:e44860,
2019
Forensic age estimation with Bayesian convolutional neural networks based on panoramic dental X-ray imaging
Forensic age estimation is the medical assessment of age in individuals for legal purposes. In current practice, medical experts use scoring-based methods to assess the most probable age and provide a minimum age estimate, which requires expensive training and is known to have poor inter-rater reliability. As an initial step towards a more objective, quantitative age estimation system, we use Bayesian convolutional neural networks to perform age and uncertainty estimation using a large data set of 12000 panoramic radiographs of the upper and lower jaws, orthopantomograms (OPTs), one of the most accurate indicators of age in subadults. Our system achieves a concordance correlation coefficient ccc=0.91 on the validation set. Importantly, our method provides quantitative estimation of prediction uncertainty, which is imperative within a legal context.
Medical Imaging with Deep Learning (MIDL),
2019
Quantification of Nematic Cell Polarity in Three-dimensional Tissues
How epithelial cells coordinate their polarity to form complex tissues and organs remains a fundamental question in biology. Here, we characterize a unique type of polarity found in liver tissue - nematic cell polarity, which is different from the common vectorial cell polarity in simple epithelia. We propose a conceptual and algorithmic framework to characterize complex patterns of polarity proteins on the surface of a cell in terms of a multipole expansion. To rigorously quantify previously observed tissue-level patterns of nematic cell polarity (Morales-Navarette et al., bioRxiv:495952, 2018), we introduce the concept of co-orientational order parameters, which generalize the known biaxial order parameters S, P, C, D from the theory of liquid crystals. Applying these concepts to three-dimensional reconstructions of single cells from high-resolution imaging data of mouse liver tissue, we show that the axes of nematic cell polarity of hepatocytes are co-aligned with the biaxially anisotropic sinusoidal network for blood transport. Thus, our study characterizes liver tissue as a biological example of a biaxial liquid crystal. The general methodology developed here could be applied to other tissues or in-vitro organoids.
arXiv,
2019
Statistical and mathematical modeling of spatiotemporal dynamics of stem cells
Statistical and mathematical modeling are crucial to describe, interpret, compare and predict the behavior of complex biological systems including the organization of hematopoietic stem and progenitor cells in the bone marrow environment. The current prominence of high-resolution and live-cell imaging data provides an unprecedented opportunity to study the spatiotemporal dynamics of these cells within their stem cell niche and learn more about aberrant, but also unperturbed, normal hematopoiesis. However, this requires careful quantitative statistical analysis of the spatial and temporal behavior of cells and the interaction with their microenvironment. Moreover, such quantification is a prerequisite for the construction of hypothesis-driven mathematical models that can provide mechanistic explanations by generating spatiotemporal dynamics that can be directly compared to experimental observations. Here, we provide a brief overview of statistical methods in analyzing spatial distribution of cells, cell motility, cell shapes and cellular genealogies. We also describe cell- based modeling formalisms that allow researchers to simulate emergent behavior in a multicellular system based on a set of hypothesized mechanisms. Together, these methods provide a quantitative workflow for the analytic and synthetic study of the spatiotemporal behavior of hematopoietic stem and progenitor cells.
In Stem Cell Mobilization (pp. 219-243). Humana, New York, NY. G. Klein, P. Wuchter (Eds.) Methods in Molecular Biology. ISSN: 1064-3745. arXiv preprint arXiv:1809.01708,
2019
Liquid-crystal organization of liver tissue
Functional tissue architecture originates by self-assembly of distinct cell types, following tissue-specific rules of cell-cell interactions. In the liver, a structural model of the lobule was pioneered by Elias in 1949. This model, however, is in contrast with the apparent random 3D arrangement of hepatocytes. Since then, no significant progress has been made to derive the organizing principles of liver tissue. To solve this outstanding problem, we computationally reconstructed 3D tissue geometry from microscopy images and analyzed it applying soft-condensed-matter-physics concepts. Surprisingly, analysis of the spatial organization of cell polarity revealed that hepatocytes are not randomly oriented but follow a long-range liquid-crystal order. This does not depend exclusively on hepatocytes receiving instructive signals by endothelial cells as generally assumed, since silencing Integrin-β1 disrupted both liquid-crystal order and organization of the sinusoidal network. Our results suggest that bi-directional communication between hepatocytes and sinusoids underlies the self-organization of liver tissue.
bioRxiv,
2018
Automated detection of the HER2 gene amplification status in Fluorescence in situ hybridization images for the diagnostics of cancer tissues
The human epidermal growth factor receptor 2 (HER2) gene amplification status is a crucial marker for evaluating clinical therapies of breast or gastric cancer. Therefore, the detection of HER2 gene amplification status is highly relevant in histopathological diagnostics. Recently, the application of convolutional neural networks (CNNs) has shown large improvements in the automation of classification and object detection in medical image analysis. Here, we propose a deep learning-based pipeline for the detection, localization and classification of interphase nuclei depending on their HER2 gene amplification state in Fluorescence in situ hybridization (FISH) images. Our pipeline combines two RetinaNet-based object localization networks which are trained (1) to detect and classify interphase nuclei into distinct classes normal, low-grade and high-grade and (2) to detect and classify FISH signals into distinct classes HER2 or centromere of chromosome 17 (CEN17). By independently classifying each nucleus twice, the two-step pipeline provides both robustness and interpretability for the automated detection of the HER2 amplification status. We demonstrate that the accuracy of this deep learning-based pipeline is on par with that of three pathologists. We demonstrate that our pipeline accurately classifies FISH images on a set of 57 validation images containing several hundreds of nuclei. Consequently, high quality FISH images can now be analyzed at once regarding their image-wide HER2 gene amplification status. The automatic pipeline is a first step towards assisting pathologists in evaluating the HER2 status of tumors using FISH images, for analyzing FISH images in retrospective studies, and for optimizing the documentation of each tumor sample by automatically annotating and reporting of the HER2 gene amplification specificities.
Scientific Reports,
2018
Hematopoietic stem cells in perisinusoidal niches are protected from aging
Experimental Hematology, 64:S43,
2018
Combining deep learning and active contours opens the way to robust, automated analysis of 3D brain cytoarchitectonics
Deep learning has thoroughly changed the field of image analysis yielding impressive results whenever enough annotated data can be gathered. While partial annotation can be very fast, manual segmentation of 3D biological structures is tedious and error-prone. Additionally, high-level shape concepts such as topology or boundary smoothness are hard if not impossible to encode in Feedforward Neural Networks. Here we present a modular strategy for the accurate segmentation of neural cell bodies from light-sheet microscopy combining mixed-scale convolutional neural networks and topology-preserving geometric deformable models. We show that the network can be trained efficiently from simple cell centroid annotations, and that the final segmentation provides accurate cell detection and smooth segmentations that do not introduce further cell splitting or merging.
Machine Learning in Medical Imaging (MLMI), LNCS 11046. bioRxiv 297689,
2018
Towards a standard exchange format for spatial, multilevel multicellular models
Standardization of model descriptions has boosted the field of systems biology over the last decade. Standard formats such as SBML and CellML have allowed the exchange of models of biochemical reaction networks between users and simulation software, enhanced reproducibility of models and enables the creation of public model repositories. However, the recent shift in systems biology towards spatial multilevel models requires new modeling formalisms and simulation software for which existing exchange formats are not suitable. Here, we discuss the major challenges in defining a standard exchange format for computational models of multilevel and multicellular systems and formulate some suggestions for the establishment of a standard exchange format for such simulation models.
Proc. Multiscale Spatial Comput. Syst. Biol.(Dagstuhl Seminar 14481),
2016
Image-based quantification and mathematical modeling of spatial heterogeneity in ESC colonies
Pluripotent embryonic stem cells (ESCs) have the potential to differentiate into cells of all three germ layers. This unique property has been extensively studied on the intracellular, transcriptional level. However, ESCs typically form clusters of cells with distinct size and shape, and establish spatial structures that are vital for the maintenance of pluripotency. Even though it is recognized that the cells’ arrangement and local interactions play a role in fate decision processes, the relations between transcriptional and spatial patterns have not yet been studied. We present a systems biology approach which combines live-cell imaging, quantitative image analysis, and multiscale, mathematical modeling of ESC growth. In particular, we develop quantitative measures of the morphology and of the spatial clustering of ESCs with different expression levels and apply them to images of both in vitro and in silico cultures. Using the same measures, we are able to compare model scenarios with different assumptions on cell–cell adhesions and intercellular feedback mechanisms directly with experimental data. Applying our methodology to microscopy images of cultured ESCs, we demonstrate that the emerging colonies are highly variable regarding both morphological and spatial fluorescence patterns. Moreover, we can show that most ESC colonies contain only one cluster of cells with high self-renewing capacity. These cells are preferentially located in the interior of a colony structure. The integrated approach combining image analysis with mathematical modeling allows us to reveal potential transcription factor related cellular and intercellular mechanisms behind the emergence of observed patterns that cannot be derived from images directly.
Cytometry Part A,
2015
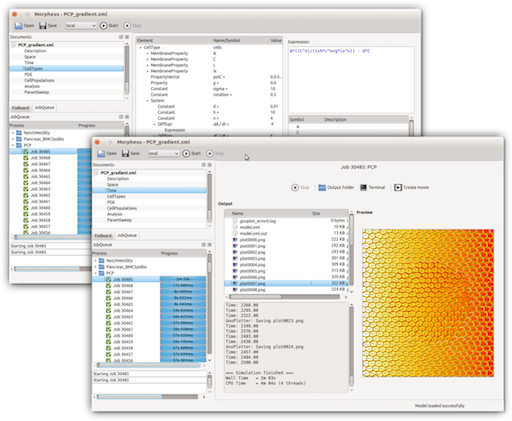
Morpheus: a user-friendly modeling environment for multiscale and multicellular systems biology
Morpheus is a modeling environment for the simulation and integration of cell-based models with ordinary differential equations and reaction-diffusion systems. It allows rapid development of multiscale models in biological terms and mathematical expressions rather than programming code. Its graphical user interface supports the entire workflow from model construction and simulation to visualization, archiving and batch processing.
Bioinformatics, 30:9,
2014
Transdifferentiation of pancreatic cells by loss of contact-mediated signaling
Replacement of dysfunctional beta-cells in the islets of Langerhans by transdifferentiation of pancreatic acinar cells has been proposed as a regenerative therapy for diabetes. Adult acinar cells spontaneously revert to a multipotent state upon tissue dissociation in vitro and can be stimulated to redifferentiate into beta-cells. Despite accumulating evidence that contact-mediated signals are involved, the mechanisms regulating acinar-to-islet cell transdifferentiation remain poorly understood.
BMC Systems Biology,
2013
On the role of lateral stabilization during early patterning in the pancreas
The cell fate decision of multi-potent pancreatic progenitor cells between the exocrine and endocrine lineages is regulated by Notch signalling, mediated by celltextendashcell interactions. However, canonical models of Notch-mediated lateral inhibition cannot explain the scattered spatial distribution of endocrine cells and the cell-type ratio in the developing pancreas. Based on evidence from acinar-to-islet cell transdifferentiation in vitro, we propose that lateral stabilization, i.e. positive feedback between adjacent progenitor cells, acts in parallel with lateral inhibition to regulate pattern formation in the pancreas. A simple mathematical model of transcriptional regulation and celltextendashcell interaction reveals the existence of multi-stability of spatial patterns whose simultaneous occurrence causes scattering of endocrine cells in the presence of noise. The scattering pattern allows for control of the endocrine-to-exocrine cell-type ratio by modulation of lateral stabilization strength. These theoretical results suggest a previously unrecognized role for lateral stabilization in lineage specification, spatial patterning and cell-type ratio control in organ development.
Journal of The Royal Society Interface,
2013