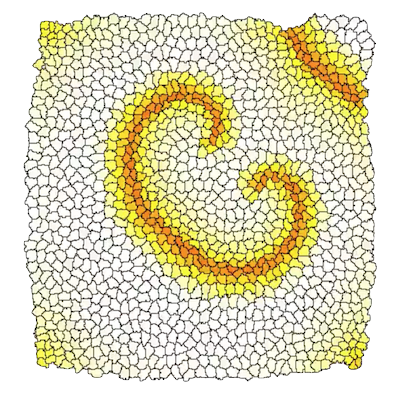
Tissue development during development, disease, and regeneration is orchestrated by a plethora of molecular pathways, cell behaviors, cell-cell signaling, and biophysics. Modeling and simulation of these processes using multi-scale cell-based model can help to unravel the interplay between these mechanisms.
Togethet with Jörn Starruß, I have developed a versatile open-source modeling and simulation environment for multicellular systems biology called Morpheus. This software tool allows rapid model development turning hypotheses in executable simulations, without requiring programming expertise. On the other hand, its flexible pligin architecture enables computational savvy experts to develop their customized tools for simulation and analysis.
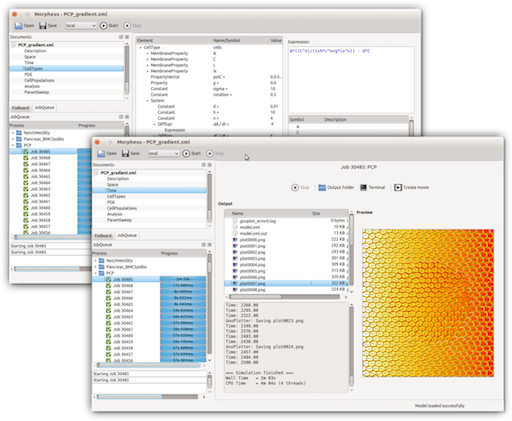
Morpheus user interface:
modeling view (back) and simulation view (front)
Morpheus allows the simulation of ordinary differential equations (ODEs) and reaction-diffusion systems (PDEs) and cell-based model cellular Potts models (CPM). It provides a comprehensive (Qt-based) GUI for model construction and simulation and uses a symbolic declarative language (MorpheusML) to specify and store simulation models. By automatic construction of a computational graph (directed acyclic graph) based on the model description, complex simulations can be correctly and efficiently executed, without the need for human specification.
Morpheus is used for research groups around the globe and is highly suitable for education in mathematical biology, systems biology and synthetic biology.
We are currently working on a number of projects to further develop Morpheus:
MultiCellML: Establishing a SBML-like model exchange format for multicellular systems biology
Software sustainability: Further establishing the community-driven development of Morpheus through flexible infrastructure, documentation and workshops.
FitMultiCell: Developing software tools for parameter inference for large-scale multicellular simulations, partnering with the developers of
pyABC.
funding
- BMBF e:Med Systems Medicine network
- DFG Research Software Sustainability programme (e-Research Technologies)
- BMBF Computational Life Science programme
collaborators
- Jörn Starruß, scientific software developer, Center for Information Services and High Performance Computing, TU Dresden
- Dr. Lutz Brusch, Center for Information Services and High Performance Computing, TU Dresden
- Prof. dr. Andreas Deutsch, Center for Information Services and High Performance Computing, TU Dresden
- Dr.-Ing. Jan Hasenauer, Institute of Computational Biology, Helmholtz Center Munich