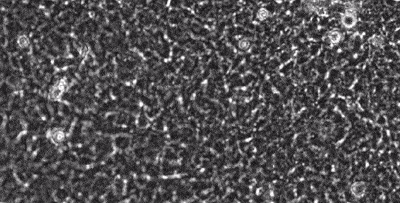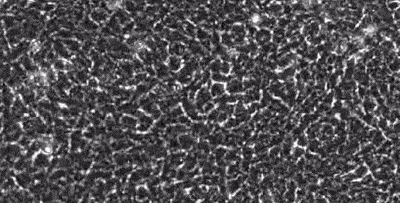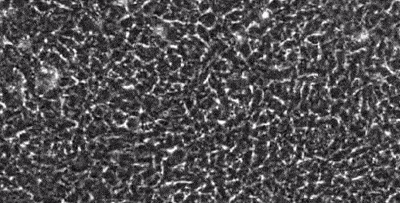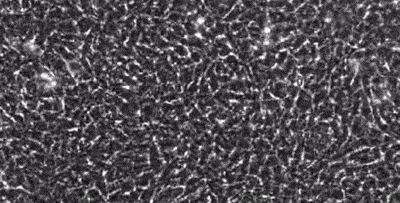
segmentation in live-cell imaging
Live-cell imaging allows the longitudinal study of the spatiotemporal behavior of cells, including their motility, their shape, their spatial interactions, their proliferation and differentiation. However, the analysis of these behaviors crucially depends on our ability to quantify them from image sequences in terms of cell segmentation and cell tracking. This is often complicated by background noise, variability in lighting and fluorescence, low signal-to-noise ratios, and e.g. the presence assay conditions such as feeding layer cells.
Together with the LOEWE Center for Cell and Gene Therapy in Frankfurt, we are developing methods to segment and track (human leukemic haematopoetic) stem cells from long time-lapse microscopy.
We use a combination of tensor decomposition methods and convolutional neural networks to segment cells in image sequences and cell tracking algorithms to construct lineage trees.
Subsequently, we employ a set of statistical methods for quantitative analysis of the differences in cell motility and proliferation to characterize different types of acute myeloid leukemia (AML).
collaborators
- Prof. dr. Michael Rieger, LOEWE Center for Cell and Gene Therapy, Department for Hematology and Oncology, University Hospital Frankfurt