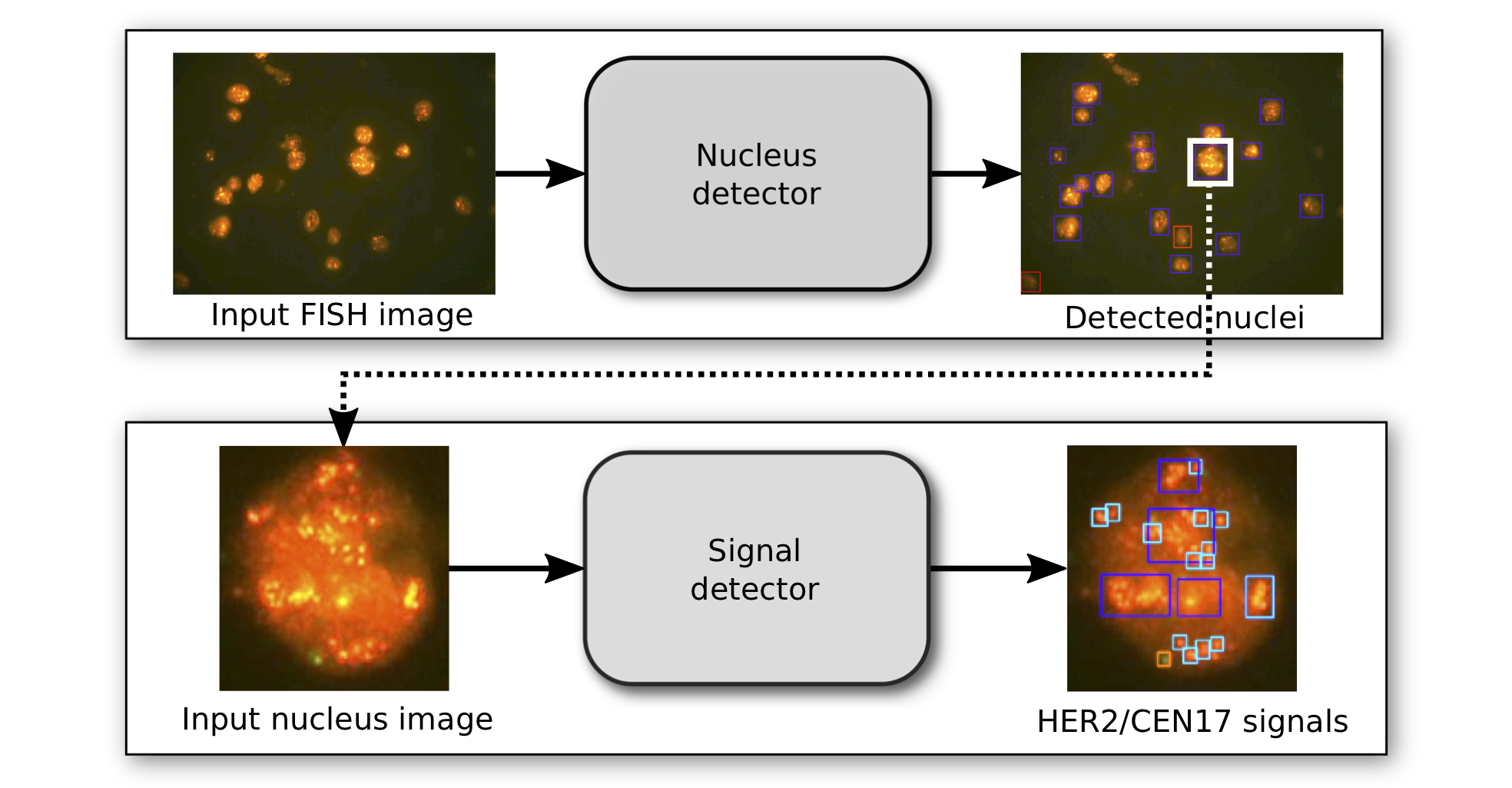
automated detection of HER2 amplification status
Detecting the HER2 overexpression or gene amplification status is crucial for treatment decisions in breast and gastric cancer. When immunohistochemistry (IHC) test results are ambiguous with respect to HER2 overexpression, the gene amplification status is tested using fluorescence in-situ hydridization (FISH). Currently, quantification of FISH tests is conducted by a trained pathologist and depends on manual counting of FISH signals (spots) in interphase nuclei and classification into low, normal of high-grade nuclei. However, this practice requires long training, is time-consuming and suffers from a high interrater variability.
Here, we automate this process using deep neural networks to localize and classify nuclei and FISH signals. In particular, we trained two independent state-of-the-art object localization networks that allow us to count and quantify the status of each nucleus and provide full medical reports on each FISH image. We demonstrate that the performance of this deep learning approach is on par with that of a team of pathologists based on nuclei-level classification Moreover, we obtain a 96% validation accuracy on the image-wide classification.
collaborators
Dr. Falk Zakrzewski, Institute for Pathology, University Hospital Dresden
Dr. Martin Weigert, Max Planck Institute for Molecular Cell Biology and Genetics (MPI-CBG) and Center for Systems Biology Dresden (CSBD).
Dr. Pia Hönscheid, Institute for Pathology, University Hospital Dresden
publications
Zakrzewski, de Back et al. Automated detection of the HER2 gene amplification status in Fluorescence in situ hybridization images for the diagnostics of cancer tissues, Scientific Reports, 2019 (to appear).
Zakrzewski, de Back et al. Automated detection of the HER2 gene amplification status in Fluorescence in situ hybridization images using deep object detection networks, Proceedings of Machine Learning Research, 2019 (to appear).