Cell shape is one of the most common cell properties used to characterize cellular phenotypes, in particular in high-content imaging and drug screening. This is due to the fact that cell morphology can be used as a proxy for a range of cellular processes including cell death, division, polarity and motility. Moreover, the shape of a cell is a relatively easy accessible property given appropriate cytoplasmic staining.
For the quantification of cell shape, various techniques are available, mostly focused on measuring classical shape features such as area, length, circularity etc., as described in our preprint.
Now, with the advent of live cell imaging techniques such as video microscopy, increasingly, data on the dynamical changes of cell shapes are also becoming accessible. However, the quantitative analysis of cell shape dynamics is much more challenging.
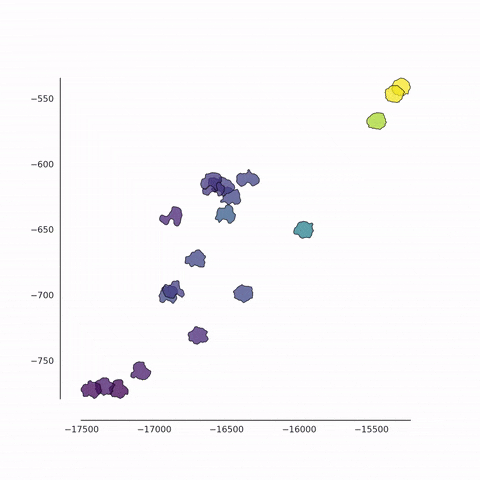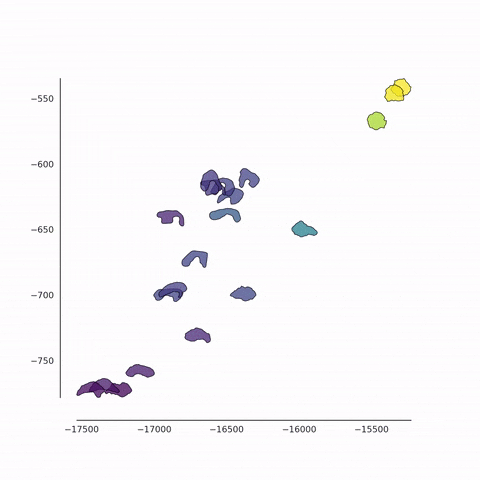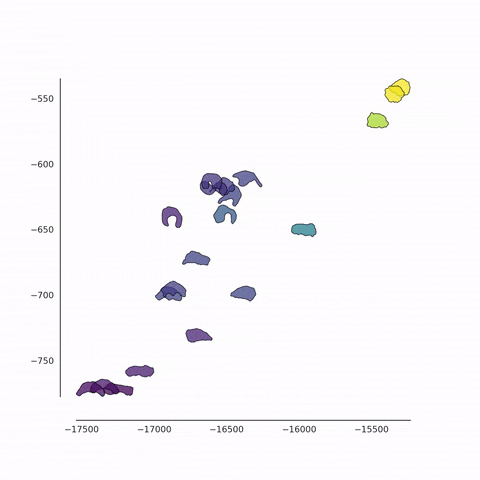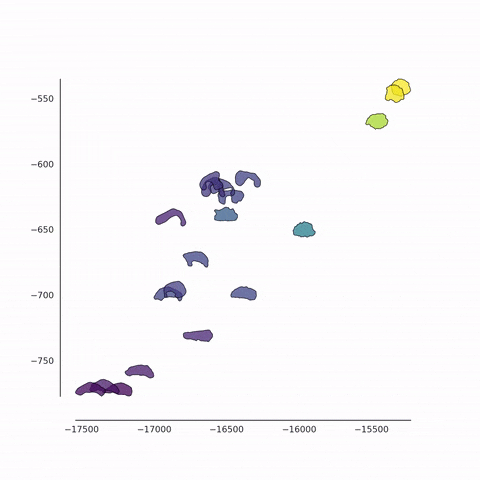
Together with Alvaro Köhn-Luque (Oslo) and Koichi Nishiyama (Kumamoto), we are developing new methods for the detection of differences between populations of cells using fluorescence video microscopy. In addition to the conventional feature-based approach, we also use a contour-based approach in which cell shapes are quantified in an unbiased fashion. These data are analysed using a variety of machine learning methods for dimensionality reduction such as PCA, t-SNE and UMAP. We also employ a set of techniques from tensor decomposition, for the multi-dimensional analysis of dynamical changes.
With Sebastian Wagner, I am developing the python package cellshapy that implements all these techniques into a convenient module. This package will be released on our github soon.
collaborators
- Sebastian Wagner, PhD student, Faculty of Medicine, TU Dresden
- Dr. Alvaro Köhn-Luque, Department of Biostatistics, Oslo University, Norway
- Dr. Koichi Nishiyama, MD, International Research Center for Medical Sciences, Kumamoto University, Japan